RESEARCH SUMMARY OF RECENT WORK
Design of DNA Targeting Molecules and Designer DNA
Professor, Nihon University
Emeritus Professor, Kyoto University Isao Saito
Pioneering Work on the Photochemistry of Biomolecules and its Application to Chemical Biology
Summary of Researches by Dr. Isao Saito
Dr. Saito is well known for his broad contributions to the photochemistry at the interface of chemistry and biology and for his outstanding leadership and influence in scientific community. His studies on photochemistry have extended over forty years with many milestones of discovery. His contribution to the photochemical science started from the pioneering work on the mechanistic study of photosensitized oxygenation of biologically important molecules in early 1970. During 1980-1993 he discovered many novel photoreactions that are important in photobiology. A prominent example is the novel reaction of photoexcited thymine base with amino acid lysine which was later shown to be extremely important in photo-cross-linking of DNA to protein in a DNA-protein complex.92,418 Saito also explored numerous novel photoreactions that find widespread applications in organic synthesis. One of such synthetically useful reactions is the general method for radical generation at the carbon attached to the hydroxyl group via a photosensitized photo-electron-transfer process.113 This method of radical generation without using organo metallic tin and silicon reagents is widely used in organic synthesis including natural product synthesis in many laboratories worldwide.
Since late 1980s his research interest was further expanded to the photochemistry of more complex biomolecules including nucleic acids. His work was aimed at gaining basic understanding of photochemistry of biomolecules and applying this understanding to the design of functionally new and practically useful molecules. Saito achieved important milestones in developing a new area of photochemistry in the interdisciplinary fields of photochemistry and chemical biology as a leader of the research project (1996-2007) of Japan Science and Technology Agency (JST). His most important contributions include, i) the discovery of new method for non-enzymatic DNA ligation using photochemistry, a breakthrough that has great implications in DNA nanotechnology and gene manipulation, ii) the pioneering work on DNA electron transfer chemistry, and iii) a new development of the most useful and cost effective gene detection technology using designed solvatochromic fluorescent nucleobases. The contributions of Saito’s work to photochemistry are chronologically summarized as bellow.
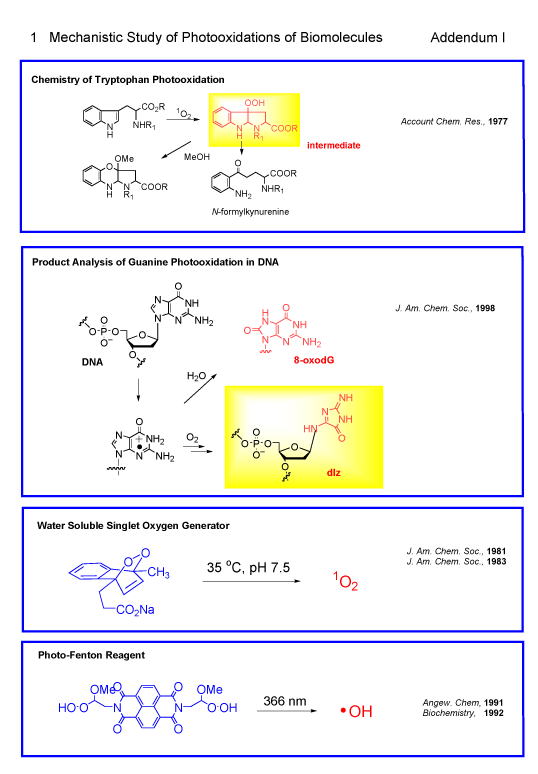
1. Mechanistic Study of Photooxidations of Biomolecules (Addendum I)417,424,426,427
During 1967-1982 Saito and Matsuura studied extensively the photosensitized oxygenations and singlet oxygen reactions of biologically important molecules such as guanine, purines, tryptophan (Trp), histidine, imidazoles, phenols and indoles.2-70 Due to their enormous efforts, the fundamental schemes of the photodynamic degradation of proteins and nucleic acids has been uncovered. Particularly, long standing problem on the mechanism of Trp photooxidation has been clarified by their extensive mechanistic study including isolation of hydroperoxide intermediates.40,48,417 Singlet oxygen reaction of electron rich double bond of enamines and indoles leading to the formation of unstable dioxetanes was experimentally verified for the first time at that time by low temperature photooxidation technique.42,52 Most importantly, Saito group developed two important reagents, water-soluble singlet oxygen generator that generates singlet oxygen without light76, 88 and photo-Fenton reagent, 157 both of which are widely used in oxidation study of biomolecules.
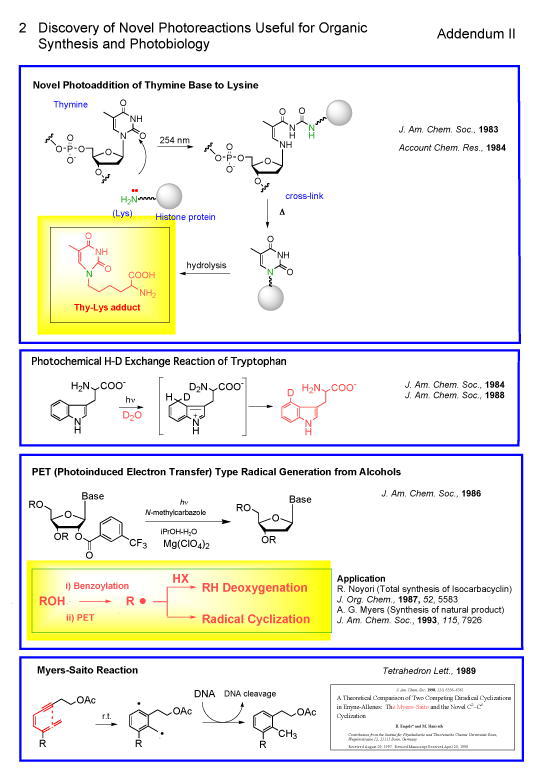
2. Discovery of Novel Photoreactions Useful for Organic Synthesis and Photobiology (Addendum II)418,432
During
1975-1990 Saito group developed numerous new photoreactions of various heterocycles
and aromatic compounds, e.g.,
synthetically useful photoadditions of aromatics66 and
heteroaromatics23,67,87 via exciplexes and novel photoisomerization
of heterocycles.37,128 The most important photoreaction in
photobiology, he discovered, was a novel photoaddition of thymine base to
lysine as described before.98,92,418
Another photobiologically important reaction was a photochemical H-D
exchange reaction at C-4 of Trp in D2O
via an internal charge transfer process
leading to quenching of Trp fluorescence.104,111
After this discovery, 126,127
many researchers extensively studied the
mechanism of the quenching of Trp
fluorescence and applied this novel photo-exchange reaction for specific
isotope labeling of Trp residue in a
protein.
The most synthetically useful photoreaction, he discovered, is the
general method for the dehydrogenation of alcohol via a photo-electron-transfer process as mentioned before.113
Another unique work of Saito’s group was the design of photo-DNA-cleaving
molecules.132,167,195 His group has explored divergent
photo-DNA-cleaving molecules which were actually used as a foot-printing agent
and for DNA sequencing.420 Saito was the inventor of a new
cyclization reaction of ene-yne-allene leading to the formation of aromatic
biradical widely known as “Myers-Saito reaction” which was later investigated
intensively from theoretical and synthetic view points by many research groups.139 He was also the first to discover
“lumiblemycin”, a photorearranged bleomycin antitumor antibiotic, which had a
higher DNA cleaving activity in the presence of light.115,132
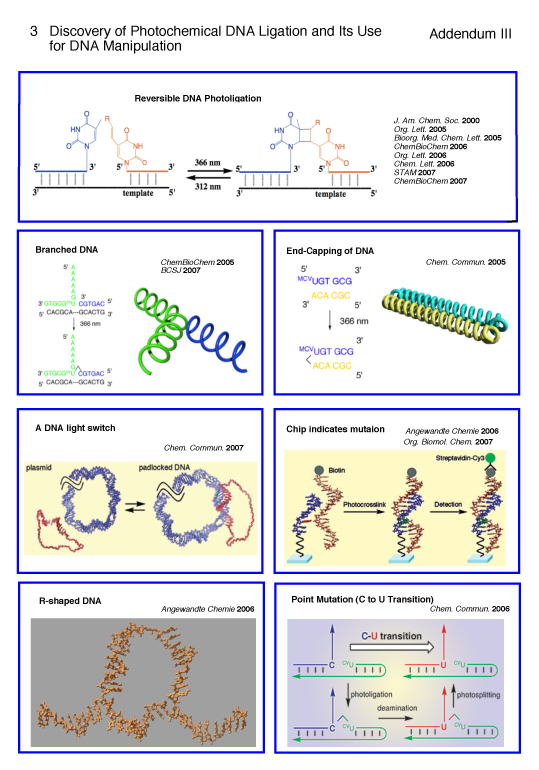
3. Discovery of Photochemical DNA Ligation and Its Use for DNA Manipulation (Addendum III)
In 2000 Saito group discovered a very important photoaddition of 5-vinyluracil derivative at the 5'-site of DNA to neighboring pyrimidine base at 3'-site of another DNA leading to the linking of two DNA chains in the presence of complementary DNA via a photoreversible [2 + 2] photocycloaddition leading to a cyclobutane formation.284,286 By utilizing this novel photochemical DNA linking reaction, he achieved an important milestone in developing a new technology for nonenzymatic template-directed DNA photoligation, a breakthrough that may have great implications in DNA nanotechnology and DNA manipulations such as photoligation of DNA chains,392 DNA end capping,393 the formation of branched and R-shaped DNAs,409 and the synthesis of many other nanoarchitechtures of DNA molecules, none of which were accessible by enzyme mediated reactions.376 Thus, the unique method is versatile and may find widespread applications for constructing DNA devices and also in recombinant DNA technology as well. This technology was further extended to the development of new method for C to U point mutation at specific site of DNA as well as to the chip-based gene detection technology. Saito and collaborators created a photoswitchable molecular padlock that prevents the access of DNA processing enzyme and could be used to block DNA-protein interaction required for the initiation of transcliption.408 These brilliant findings opened a new possibility for the application of photochemistry to gene technology and molecular biology.
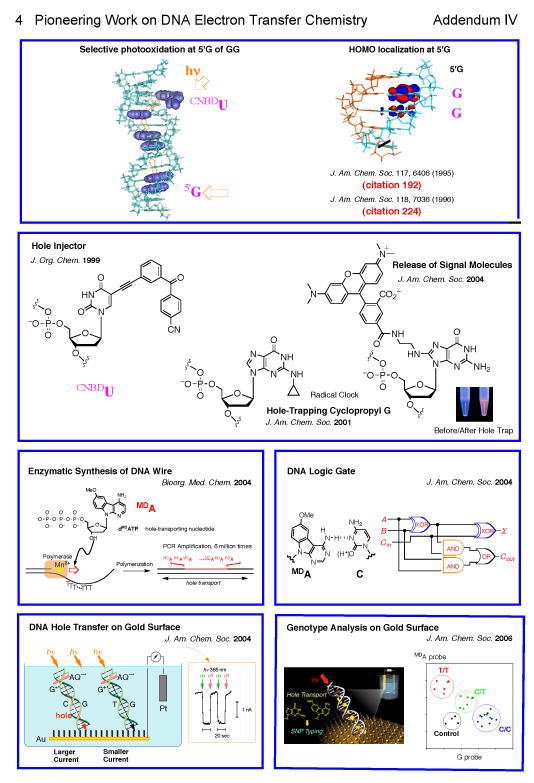
4. Pioneering Work on DNA Electron Transfer Chemistry (Addendum IV)421,436
In 1991 Saito and coworkers have demonstrated for the first time that photoirradiation of duplex DNA in the presence of an electron accepting photosensitizer resulted in a highly selective cleavage at 5'-site of GG doublet via a photoinduced electron transfer process.157,195 During 1995-1996 his group published two important papers demonstrating that 5'- site of GG doublet is the most electron-donating site in DNA and the hole generated by photo-electron transfer is trapped preferentially at this site (JACS, 117, 6406, 1995; JACS 118, 7063, 1996). Since this important finding, an unprecedented flurry of activities had started aiming to understand the mechanism of charge transport through DNA duplex which is still continuing all over the world. Actually, these two papers were ranked with world top 100 of high-impact papers most frequently cited (citation 416) from 1995 to 2001 (data from ISI). Saito group421 has greatly contributed to the elucidation of the charge transport mechanism through DNA by exploiting newly designed tools such as a novel hole-injector,270 hole-trapping cyclopropyl-guanosine,305 and dye-releasing hole-trap.357 Also important was Saito’s development of the design of artificial nucleobase MDA which possessed a very low oxidation potential but was not destroyed by photoirradiation, unlike photolabile guanine base.334 DNA duplex containing consecutive MDA was shown to behave like a DNA wire.334,372 He also demonstrated that DNA hole transport chemistry is applicable to genotyping on Au surface273,394 and usable as a DNA logic gate.364
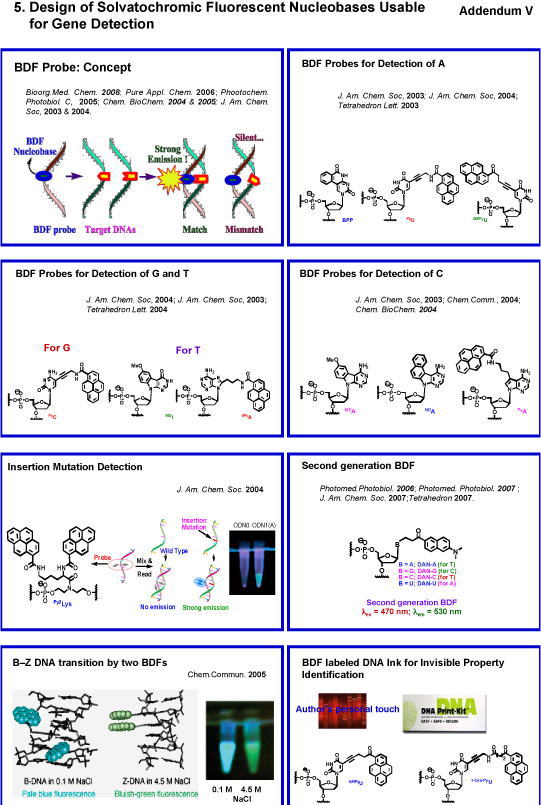
5. Design of Solvatochromic Fluorescent Nucleobases for Gene Detection (Addendum V)422
The design of intelligent solvatochromic fluorescent molecules is very important in basic photochemistry as well as for the application as a fluorescence sensor. Saito group devised conceptually new fluorescent nucleobases that can discriminate the opposite base on a complementary strand by attaching a newly designed solvatofluorochromic chromophore to natural nucleobases.335,341.359 Oligonucleotides containing such intelligent fluorescent nucleobases were named as base-discriminating fluorescent (BDF) probe and used for gene detection. Saito was the pioneer who first succeeded in the design of BDF bases which recognize four different bases (A, T, G, and C) effectively and used these BDF bases for SNP genotyping.359, 404,410 His inventions of BDF bases have extended to the discovery of the FRET based strategy for genotyping406 and pyrene excimer emission based detection of insertion/deletion polymorphism.360 In collaboration with industry, DNA chip containing BDF–package array was developed and now commercially available. Since Saito’s unique work was published in 2004, numerous papers on fluorescence DNA probe have appeared thereafter. In a recent new project Saito group has just embarked on the exciting area of DNA ink technology by exploiting BDF probe of unique fluorescence property that is utilized in invisible protection for personal property and in crime prevention as well as in detection techniques. The pioneering work on BDF base by Saito group has stimulated many researchers to develop intelligent fluorescent molecules for gene detection, which is now actively ongoing all over the world.
Saito has received numerous awards in recognition of his scientific
achievement, which include i) Japan Chemical Society Award for Distinguished
Young Chemists, ii) Japan Photochemistry Association Prize, iii) Naito
Foundation Award, iv) The Japan Chemical Society Prize (Highest Award of JCS)
and v) Japan Photochemistry Association Honorary Award. He is named as a Fellow
of the American Association for the Advancement of Science (AAAS) and as IUPAC
fellow. He was appointed to the leader of the research projects CREST
(1996-2001) and SORST (2002-2007) of Japan Science and Technology Agency (JST).
He served as a councilor, an executive committee or review board member in
numerous domestic and international societies including Japan Photochemistry
Association and American Society for Photobiology. He has been serving as
Advisory and Editorial Board member of Bioorganic
Chemistry (Academic Press) (1990-present), J. Photochem. Photobiol. A: Chemistry (Elsevier) (2000-present), J. Photochem. Photobiol., C; Reviews
(2000-present), and Photomedicine &
Photobiology (1995-present).
His contributions to IUPAC Symposium on Photochemistry are as follows;
i) IUPAC Titular Member (Organic Division, Photochemistry, 1994-1998), ii)
Plenary Lecturer (1992) and Invited Lecturer (2006), iii) Symposium Organizing
Committee (1990), and iv) International Foundation of Photochemistry, trustee
(1998-2005).
He is the author and coauthor of 416
original papers, 38 review articles and 35 books (including reviews and books
written in Japanese) and the holder of 40 patents.
The incorporation of designer nucelobases
into DNA has become important in molecular biology, genetics, molecular
medicine and DNA microarray technology. For the past several years we designed
different types of intriguing DNA targeting molecules that are useful for
chemical genomics and molecular biology. The aim of this research project is to
design new DNA targeting molecules and intelligent nucleobases that are useful
for molecular biology and genome industry. These include, 1) deisign and
synthesis of tailor-made DNA alkylating agents possessing anticancer activity,
2) molecular design of mismatch recognition molecules , 3)
development of new methods for photochemical DNA manipulation, 4)
mechanistic study of DNA-mediated hole transport, 5) design of artificial DNA nano-wire, and 6)
development of a new methodology for
mapping DNA HOMO. The goal of this research is to devise new
methodologies and nano-materials which are particularly useful for chemical
genomics.
6. Design of Tailor-Made Antitumor Agents
Sequence-specific DNA recognition in the minor groove depends on the sequence of side-by-side aromatic amino acid pairing oriented in the amino-carboxyl (N-C) direction with respect to the 5'-3' direction of the DNA helix. Antiparallel pairing of Im opposite Py (Im/Py) recognizes a G-C base pair, whereas a Py/Py pair recognizes A-T or T-A base pairs. These Py-Im hairpin polyamides have binding affinity and sequence-specificity comparable to those of transcription factors. Therefore, Py-Im hairpin polyamides can potentially control DNA replication and gene expression. We recently demonstrated that a conjugate between segment A of duocarmycin A (Du) and a Py-Im hairpin polyamide selectively alkylates one of the matched sequences within a 450-bp DNA fragment. Sequence-specific alkylation was observed even at 50 nanomolar concentrations of the alkylating agent.1 However, the alkylation in this system was rather slow and inefficient. In trying to improve the alkylating activity of the polyamide, we found that insertion of a vinyl linker (L) between the polyamide and cyclopropylpyrroloindole (CPI) adjusted the location of the reactive cyclopropane ring, enhancing its reactivity. As a result using ImPyLDu86, highly sequence-specific cooperative double-strand alkylation of DNA is facilitated through highly cooperative homodimer formation. Quantitative analysis indicated that the efficiency of this system reached 69%.2
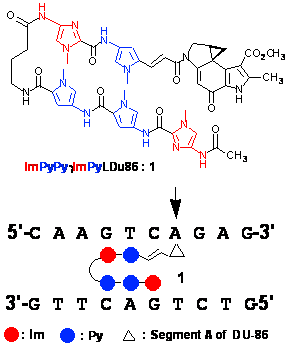
Figure 1. Chemical structure of compound 1 and a schematic representation of the recognition of the AGTCAG sequence by 1. Arrows indicate the site of alkylation.
New hairpin polyamide–CPI conjugate, compound 1, was synthesized and their DNA alkylating activities compared with the previously prepared hairpin conjugate conjugate by high-resolution denaturing gel electrophoresis using 450-bp DNA fragments and by HPLC product analysis of the synthetic decanucleotide.3 Hairpin CPI conjugate 1 alkylated the A of 5’-(A/T)G(A/T)CA-3’ efficiently at nanomolar concentrations. The significantly improved reactivity of the alkylating hairpin polyamides 1 was further confirmed by HPLC product analysis using a synthetic decanucleotide. The results suggest that hairpin polyamide–CPI conjugate 1 alkylates effectively according to Py-Im pairing rule, and with a new mode of recognition in which the Im-vinyl linker (L) pair targets G-C base pairs. These results demonstrate that incorporation of the vinyl linker pairing with Im dramatically improves the reactivity of hairpin polyamide–CPI conjugates. Examination of antitumor activity using a panel of 39 human cancer cell lines demonstrates that 1 has remarkably strong and unique biological activity against several cell lines. This paves the way for “tailor-made antitumor agent” by sequence-specific alkylating Py-Im polyamides.
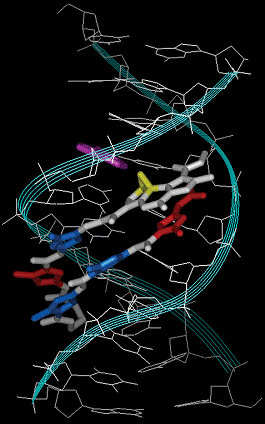
Figure 2. Energy-minimized structure of
the d(CAAGTCAGAG)/ d(CTCTGACTTG)-1
complex. Each strand of DNA is drawn in white and ribbon representation is
drawn in light blue. Py and Im are drawn
in blue and red, respectively. The
reacting A base is drawn in purple. The
cyclopropane units in the Du moiety of the hybrids are drawn in yellow.
7. G-G Mismatch Recognition
To develop a conceptually new method for rapid discovery and detection of single nucleotide polymorphisms (SNPs), we have succeeded for the first time in designing and synthesizing ligands that specifically bind with high affinity (Kd = 53 nM) to the guanine (G)-guanine mismatch, one of four SNP types. The G-G mismatch would be significantly stabilized when two naphthyridines simultaneously bind to both G bases within the base-stacks of the DNA duplex by forming 6 hydrogen bonds (Figure 3).4 Detection of the G-G mismatch was performed by a surface plasmon resonance (SPR) assay using a sensor chip carrying the G-G specific ligand on its surface. The accuracy of the G-G mismatch detection by the SPR sensor was demonstrated by a marked SPR response obtained only for the DNA containing the G-G mismatch. DNAs containing G-A and G-T mismatches, as well as a fully matched duplex, produced only a weak response.
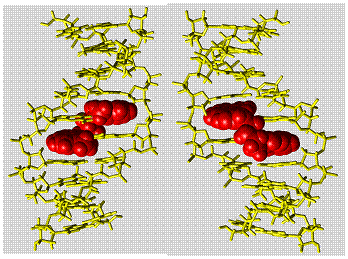
Figure 3. Molecular modeling simulations for the
complex of G-G mismatch DNA and naphthyridine dimer.
8. Photochemical DNA Manipulation
We developed a new method for reversible photochemical DNA ligation by using 5-vinyluracil derivatives that were incorporated into 5’ end of oligonucleotides in the presence of template DNA.5 By using this reversible DNA photoligation, we were able to prepare circular DNA, branched DNA, DNA-PNA chimera and capped DNA.6 We also demonstrated a new method for C to U transition by means of this new DNA manipulation technology. Photopadlocking of plasmid DNA has also been accomplished by this photoligation technique.
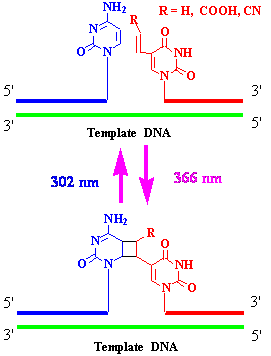
Figure 4. Photoinduced reversible DNA ligation using 5-vinyluracil at 5' end in the presence of template DNA.
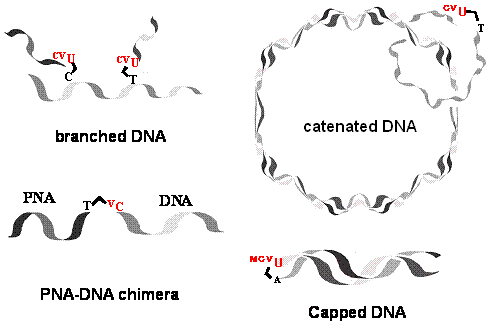
Figure 5. Synthesis of branced DNA, catenated DNA, PNA-DNA chimera and capped DNA by photoligation technique.
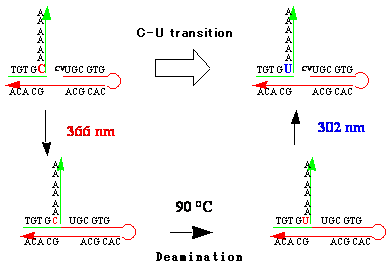
Fiigure 6. Point mutation from C to U by photoligation technique.
9. DNA Mediated Hole Transport and Ultimate DNA Wire
Guanine radical cation (hole) produced by one-electron oxidation of DNA is known to migrate to remote GG sites through DNA π-stack. We demonstrated such hole transport through DNA is modulated by binding to BamHI restriction enzyme.7 Since hole migration through DNA π-stack always competes with hole trapping with water and/or oxygen, destruction of guanine base is inevitable during charge transport process through natural DNA. Based on these basic studies on DNA mediate hole transport, we finally succeeded to design a new nucleobase, methoxybenzodeazaadenine (MDA) that has a stronger hole transport ability than natural guanine and is not destroyable during hole transport process. . By incorporating MDAs in duplex DNA, we have demonstrated a protocol for real DNA nano-wire that is quite stable and has a higher charge transport ability than natural DNA.9
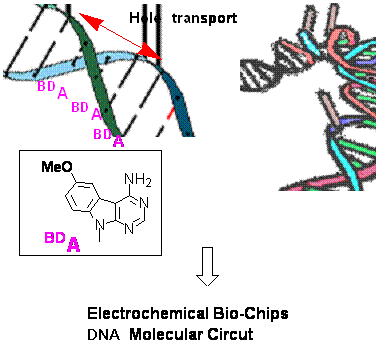
Figure 7. Ultimate DNA nanowire containing BDA as a base.
10. DNA HOMO Mapping
The highest occupied molecular orbital (HOMO) of organic molecules plays an important role in chemical reactions by interacting with LUMO of reactant molecules. We proposed an experimental method for mapping HOMO of B-DNA using Co(II) ion and benzoyl peroxide in combination with ab initio MO calculations of G-rich sequences and suggested an important but unestablished binding force, the interaction of the HOMO of duplex DNA with LUMO of DNA binders such as metal ions, drugs and proteins.8 We then examined the interaction of G runs with Mn(II) and Co(II) ions by means of 15N NMR using ODNs containing 15N-enriched guanine at N7 in GG and GGG sequences. As increasing the concentration of Mn(II), broadening of the signals of 5’G of GG and the middle G of GGG occurred selectively. These selectivities were in good agreement with HOMO distributions in G runs obtained by recent high level theoretical calculations. The interaction of electron deficient metal ions to the N7 of electron-rich G in G runs is likely a HOMO-controlled process, and as a consequence, the selectivity of the G-metal ion interactions obtained by this 15N -NMR studies would directly reflect the HOMO distribution in DNA.
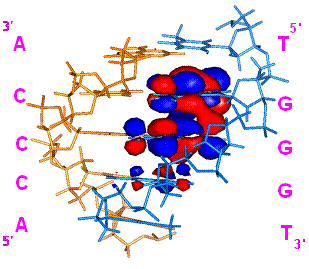
Figure 8. DNA HOMO mapping by metal ion binding as studied by 15N NMR.
References
1) Z.-F. Tao, T. Fujiwara, I. Saito, H. Sugiyama, J. Am. Chem. Soc. 1999, 121, 4961-4967
2) Z.-F. Tao, I. Saito, H. Sugiyama, J. Am. Chem. Soc. 2000, 122, 1602-1608.
3) Bando, T.; Narita, A.; Saito, I.; Sugiyama, H. Chem. Eur. J. 2002, 8, 4781-4790.
4) Nakatani, K.; Sando, S.; Saito, I Nat. Biotechnol. 2001, 19, 51-55.
5) K. Fujimoto, S. Matsuda, N. Takahashi, I. Saito, J. Am. Chem. Soc. 2000, 122, 5893.
6) K. Fujimoto, N. Ogawa, M. Hayashi, S. Matsuda, I. Saito, Tetrahedron Lett. 2000, 41,9347.
7) K. Nakatani, C. Dohno, A. Ogawa, I. Saito, Chem. Biol. 2002, 9, 361.
8) I. Saito, T. Nakamura, K. Nakatani, J. Am. Chem. Soc. 2000, 122, 3001.
9) A. Okamoto, K. Kanatani, T. Taiji, I. Saito, J. Am. Chem. Soc. 2003, 125, 1172-1173.